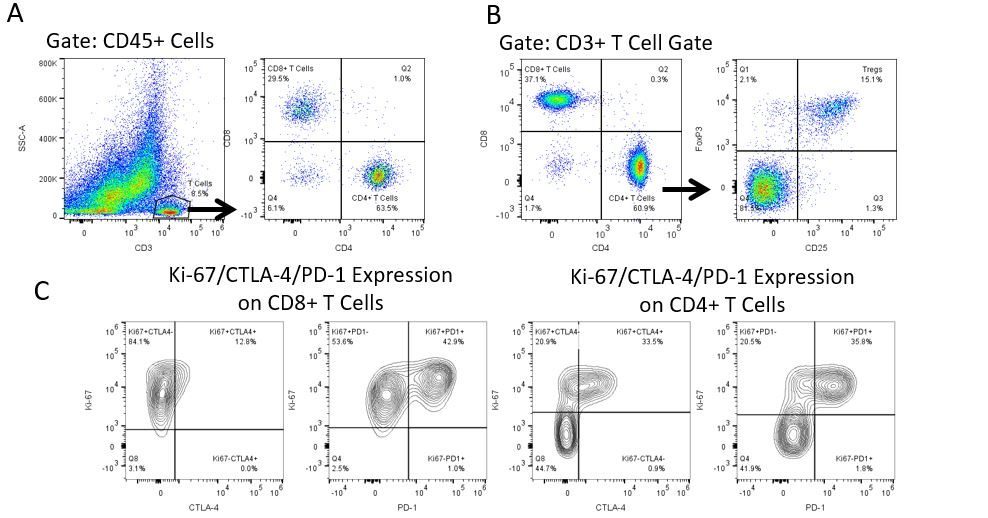
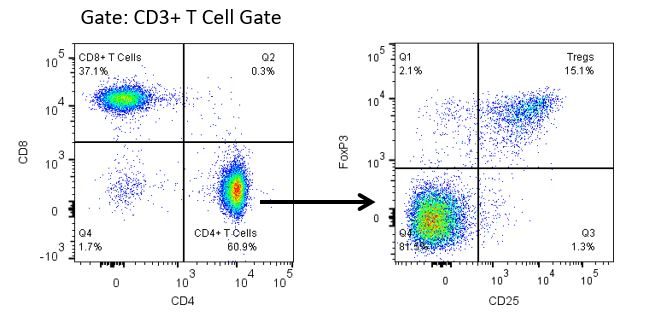
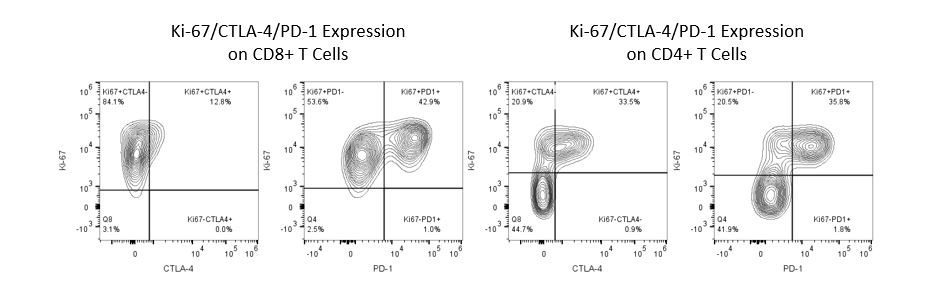
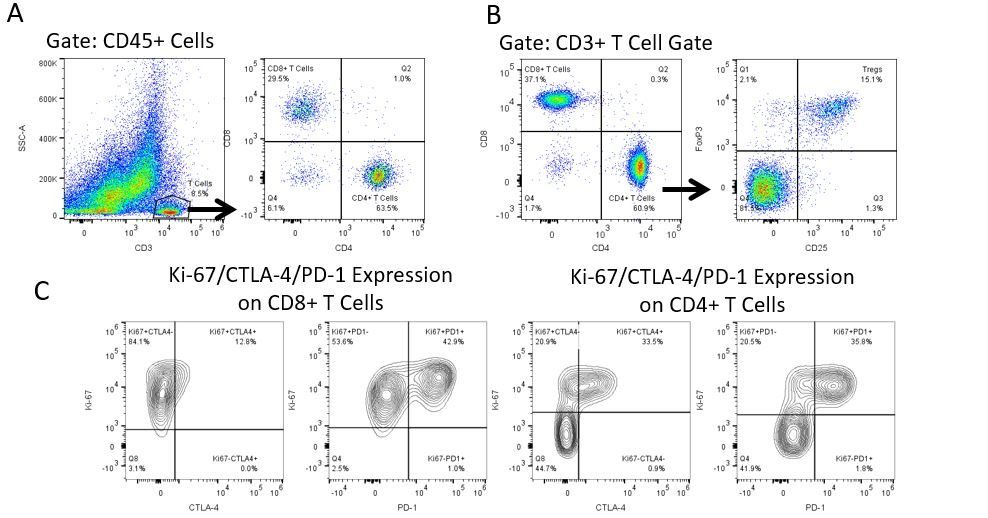
癌症研究和治疗中最近的最新突破一直在免疫肿瘤学领域,利用新型免疫治疗宿主的免疫系统以更有效地靶向并破坏肿瘤细胞。在具有免疫调节活动的FDA批准的治疗药物中是抗体和细胞因子的免疫治疗,小分子药物和基于细胞的疗法。这些和其他疗法在延长生存时的成功使得重点是在治疗癌症时具有更高疗效的新单药和组合疗法的发展。
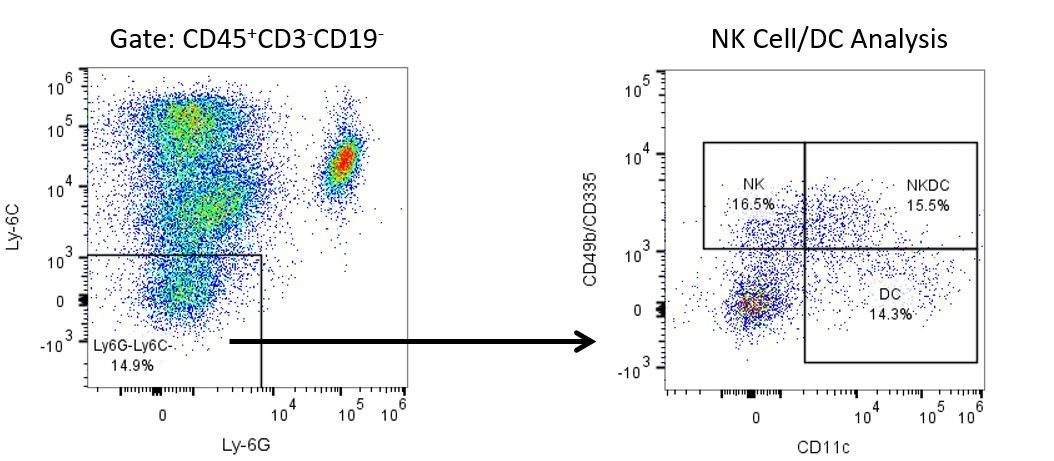
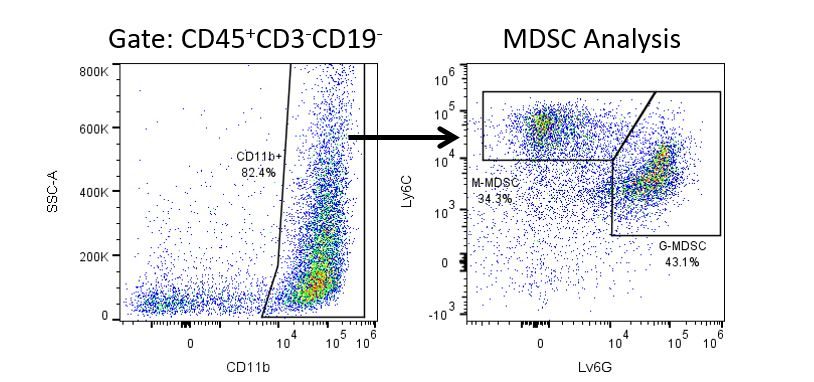
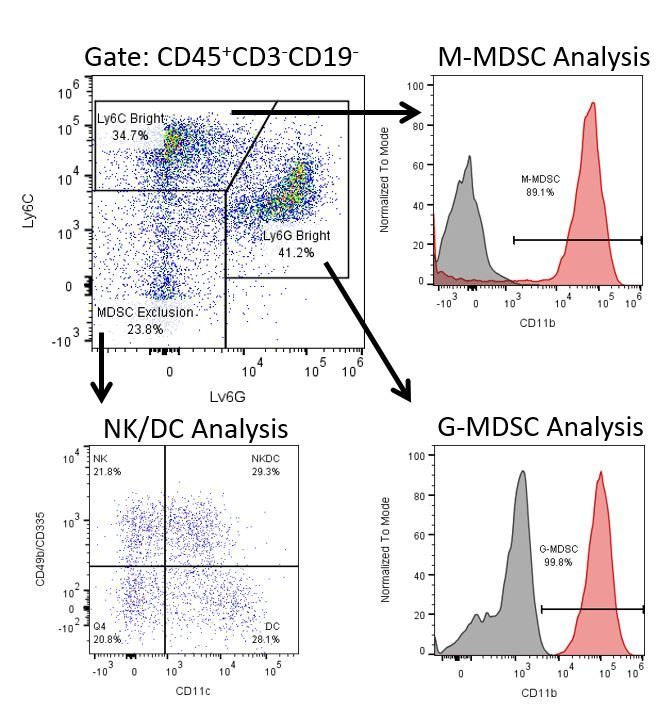
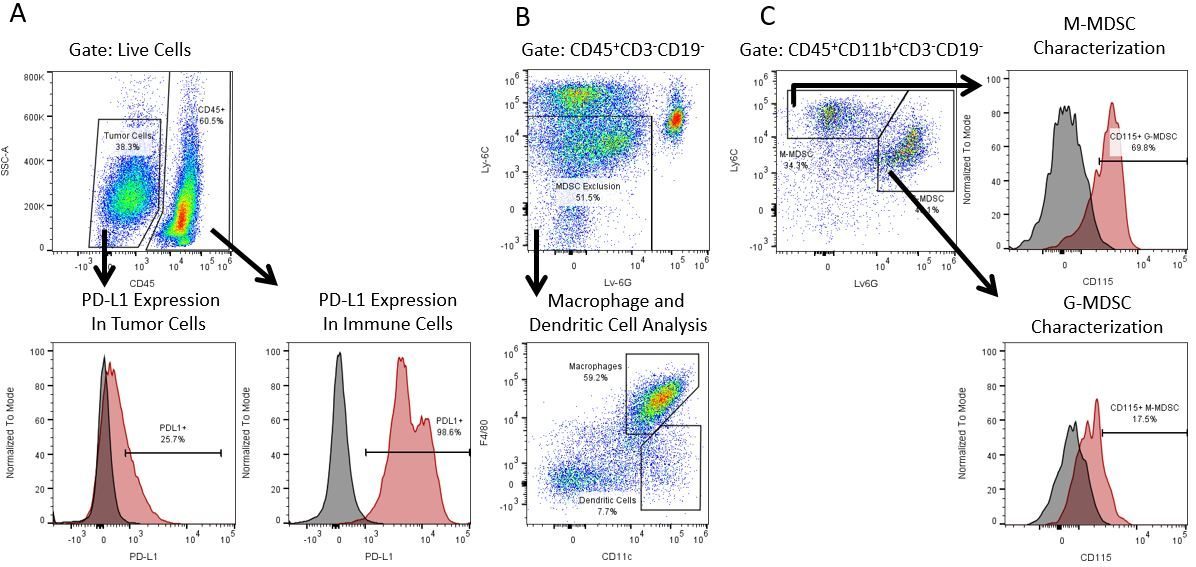
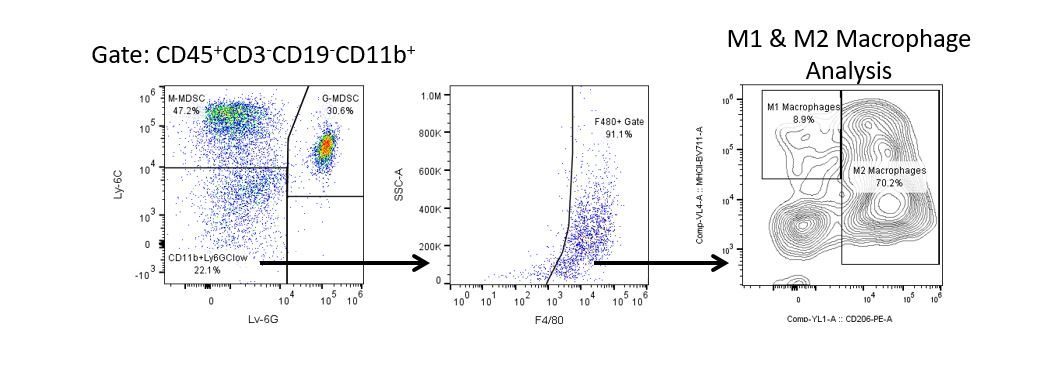
免疫疗法的生产开发需要高通量和对肿瘤微环境,外周血和其他宿主组织的免疫系统的鲁棒定量分析。
为了满足这一需求,我们的合同流式细胞术服务,为您提供先进的、最先进的分析流式细胞术资源,以支持您的药物开发需求的所有方面。与我们一起进行您的样本生成研究,或将您的临床前或非clia监管的临床样本连夜运送给我们,我们将为您处理流式细胞术。yaboapp体育官网